Supplemental Material 4
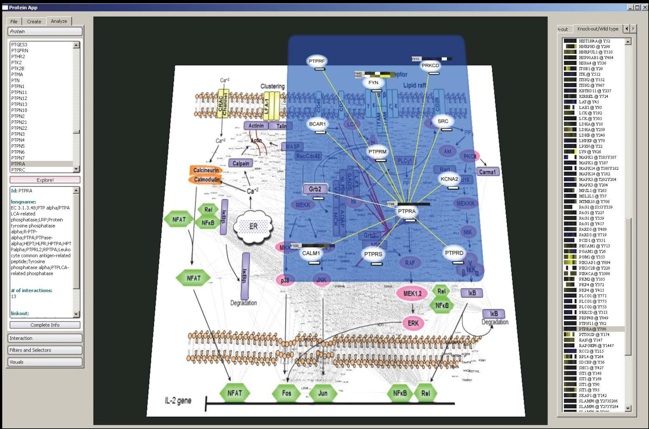
Supplemental Material 4: Custom-made exploration software for navigation through protein-protein interactions in the context of the TCR signaling pathway. This software is useful in the manual exploration of protein interaction networks, canonical signaling pathways, and quantitative proteomic data. Proteins and their interactions are imported into the software from the HPRD protein interaction database and structured according to the canonical TCR signaling pathway. Heatmaps representing quantitative data collected from phosphoproteomic comparative analysis experiments are shown both in the main image and in the frame on the right. Clicking on a protein in the main image or in the list on the right re-centers that protein in a semi-transparent overlay, displaying that protein on top of the main image of the TCR signaling pathway. Systematic navigation of this pathway can be accomplished by crawling through the network, revealing both a protein’s local known interactions and its orientation within the global network. Meta information regarding the protein of interest, such as interactions, filters, highlighters, and other operators, are also available in a panel on the left.